Fits a Bayesian spatial linear model with spatial process parameters and the noise-to-spatial variance ratio fixed to a value supplied by the user. The output contains posterior samples of the fixed effects, variance parameter, spatial random effects and, if required, leave-one-out predictive densities.
Usage
spLMexact(
formula,
data = parent.frame(),
coords,
cor.fn,
priors,
spParams,
noise_sp_ratio,
n.samples,
loopd = FALSE,
loopd.method = "exact",
verbose = TRUE,
...
)Arguments
- formula
a symbolic description of the regression model to be fit. See example below.
- data
an optional data frame containing the variables in the model. If not found in
data, the variables are taken fromenvironment(formula), typically the environment from whichspLMexactis called.- coords
an \(n \times 2\) matrix of the observation coordinates in \(\mathbb{R}^2\) (e.g., easting and northing).
- cor.fn
a quoted keyword that specifies the correlation function used to model the spatial dependence structure among the observations. Supported covariance model key words are:
'exponential'and'matern'. See below for details.- priors
a list with each tag corresponding to a parameter name and containing prior details.
- spParams
fixed value of spatial process parameters.
- noise_sp_ratio
noise-to-spatial variance ratio.
- n.samples
number of posterior samples to be generated.
- loopd
logical. If
loopd=TRUE, returns leave-one-out predictive densities, using method as given byloopd.method. Default isFALSE.- loopd.method
character. Ignored if
loopd=FALSE. Ifloopd=TRUE, valid inputs are'exact'and'PSIS'. The option'exact'corresponds to exact leave-one-out predictive densities which requires computation almost equivalent to fitting the model \(n\) times. The option'PSIS'is faster and finds approximate leave-one-out predictive densities using Pareto-smoothed importance sampling (Gelman et al. 2024).- verbose
logical. If
verbose = TRUE, prints model description.- ...
currently no additional argument.
Value
An object of class spLMexact, which is a list with the
following tags -
- samples
a list of length 3, containing posterior samples of fixed effects (
beta), variance parameter (sigmaSq), spatial effects (z).- loopd
If
loopd=TRUE, contains leave-one-out predictive densities.- model.params
Values of the fixed parameters that includes
phi(spatial decay),nu(spatial smoothness) andnoise_sp_ratio(noise-to-spatial variance ratio).
The return object might include additional data used for subsequent prediction and/or model fit evaluation.
Details
Suppose \(\chi = (s_1, \ldots, s_n)\) denotes the \(n\) spatial locations the response \(y\) is observed. With this function, we fit a conjugate Bayesian hierarchical spatial model $$ \begin{aligned} y \mid z, \beta, \sigma^2 &\sim N(X\beta + z, \delta^2 \sigma^2 I_n), \quad z \mid \sigma^2 \sim N(0, \sigma^2 R(\chi; \phi, \nu)), \\ \beta \mid \sigma^2 &\sim N(\mu_\beta, \sigma^2 V_\beta), \quad \sigma^2 \sim \mathrm{IG}(a_\sigma, b_\sigma) \end{aligned} $$ where we fix the spatial process parameters \(\phi\) and \(\nu\), the noise-to-spatial variance ratio \(\delta^2\) and the hyperparameters \(\mu_\beta\), \(V_\beta\), \(a_\sigma\) and \(b_\sigma\). We utilize a composition sampling strategy to sample the model parameters from their joint posterior distribution which can be written as $$ p(\sigma^2, \beta, z \mid y) = p(\sigma^2 \mid y) \times p(\beta \mid \sigma^2, y) \times p(z \mid \beta, \sigma^2, y). $$ We proceed by first sampling \(\sigma^2\) from its marginal posterior, then given the samples of \(\sigma^2\), we sample \(\beta\) and subsequently, we sample \(z\) conditioned on the posterior samples of \(\beta\) and \(\sigma^2\) (Banerjee 2020).
References
Banerjee S (2020). "Modeling massive spatial datasets using a conjugate Bayesian linear modeling framework." Spatial Statistics, 37, 100417. ISSN 2211-6753. doi:10.1016/j.spasta.2020.100417 .
Vehtari A, Simpson D, Gelman A, Yao Y, Gabry J (2024). "Pareto Smoothed Importance Sampling." Journal of Machine Learning Research, 25(72), 1-58. URL https://jmlr.org/papers/v25/19-556.html.
Author
Soumyakanti Pan span18@ucla.edu,
Sudipto Banerjee sudipto@ucla.edu
Examples
# load data
data(simGaussian)
dat <- simGaussian[1:100, ]
# setup prior list
muBeta <- c(0, 0)
VBeta <- cbind(c(1.0, 0.0), c(0.0, 1.0))
sigmaSqIGa <- 2
sigmaSqIGb <- 0.1
prior_list <- list(beta.norm = list(muBeta, VBeta),
sigma.sq.ig = c(sigmaSqIGa, sigmaSqIGb))
# supply fixed values of model parameters
phi0 <- 3
nu0 <- 0.75
noise.sp.ratio <- 0.8
mod1 <- spLMexact(y ~ x1, data = dat,
coords = as.matrix(dat[, c("s1", "s2")]),
cor.fn = "matern",
priors = prior_list,
spParams = list(phi = phi0, nu = nu0),
noise_sp_ratio = noise.sp.ratio,
n.samples = 100,
loopd = TRUE, loopd.method = "exact")
#> ----------------------------------------
#> Model description
#> ----------------------------------------
#> Model fit with 100 observations.
#>
#> Number of covariates 2 (including intercept).
#>
#> Using the matern spatial correlation function.
#>
#> Priors:
#> beta: Gaussian
#> mu: 0.00 0.00
#> cov:
#> 1.00 0.00
#> 0.00 1.00
#>
#> sigma.sq: Inverse-Gamma
#> shape = 2.00, scale = 0.10.
#>
#> Spatial process parameters:
#> phi = 3.00, and, nu = 0.75.
#> Noise-to-spatial variance ratio = 0.80.
#>
#> Number of posterior samples = 100.
#>
#> LOO-PD calculation method = exact.
#> ----------------------------------------
beta.post <- mod1$samples$beta
z.post.median <- apply(mod1$samples$z, 1, median)
dat$z.post.median <- z.post.median
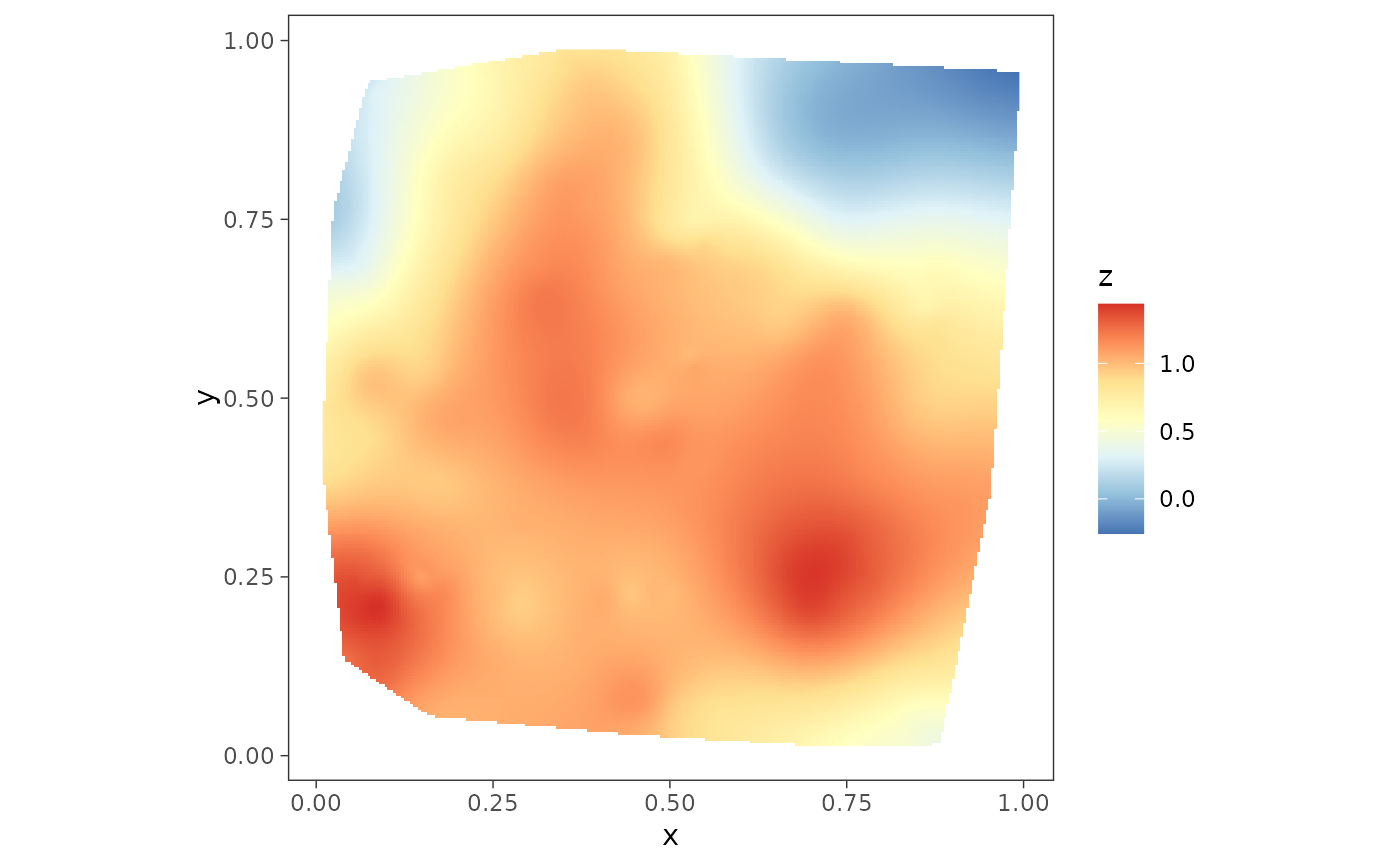
plot1 <- surfaceplot(dat, coords_name = c("s1", "s2"),
var_name = "z_true")
plot2 <- surfaceplot(dat, coords_name = c("s1", "s2"),
var_name = "z.post.median")
plot1
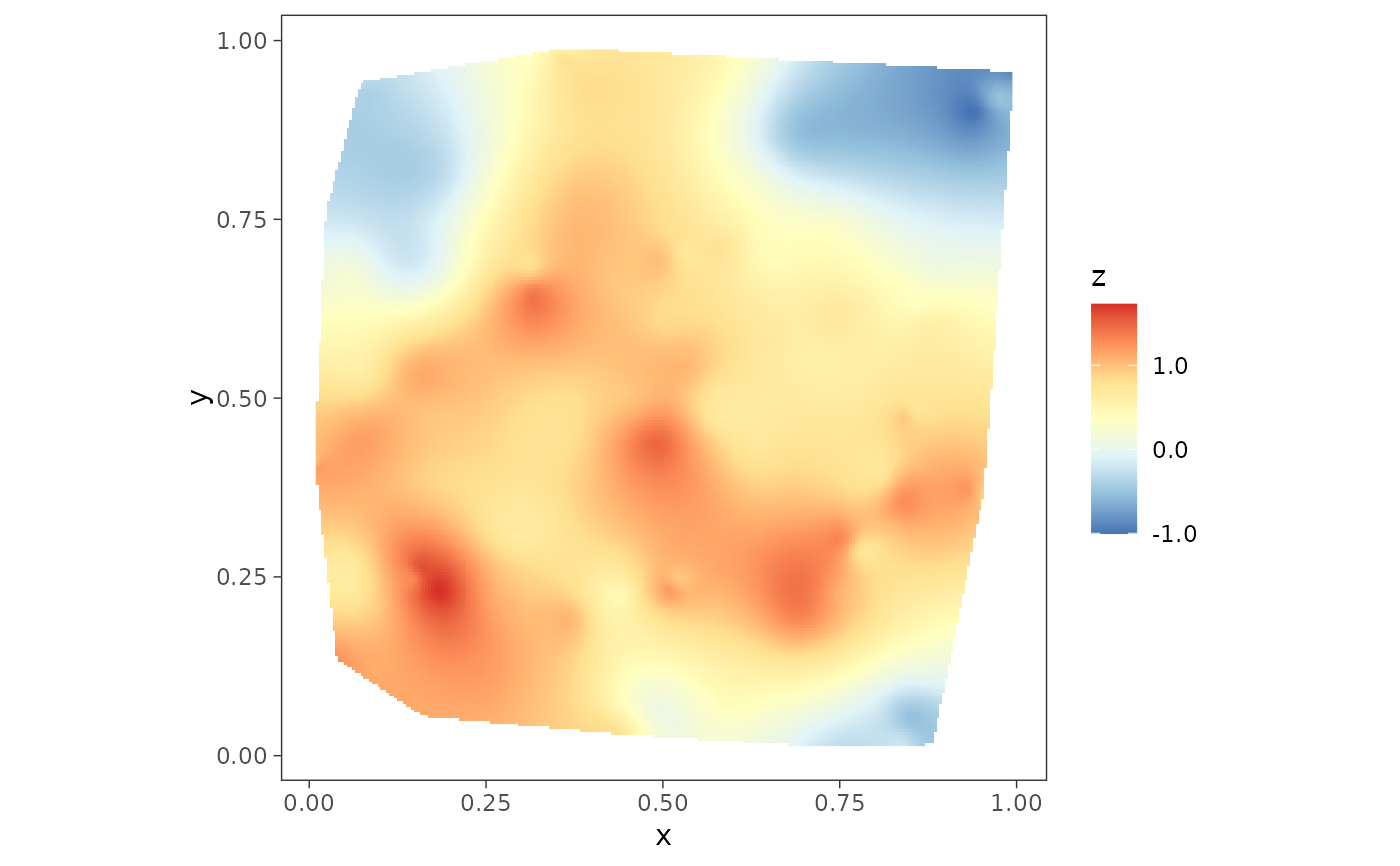 plot2
plot2